A recent tweet:
made me think (1) has it really been 5 years, (2) gee, my ggplot skills were dreadful back then and (3) did I really not know how to correct for the increase in total publications?So here is the update, at Github and the report.
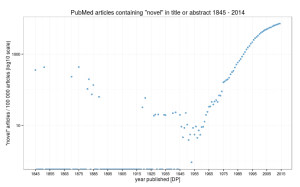
“Novel” findings, as judged by the usage of that word in titles and abstracts really have undergone a startling increase since about 1975. Indeed, almost 7.2% of findings were “novel” in 2014, compared with 3.2% for the period 1845 – 2014. That said, if we plot using a log scale as suggested by Tal on the original post, the rate of usage appears to be slowing down. See image, right (click for larger version).
As before, none of this is novel.